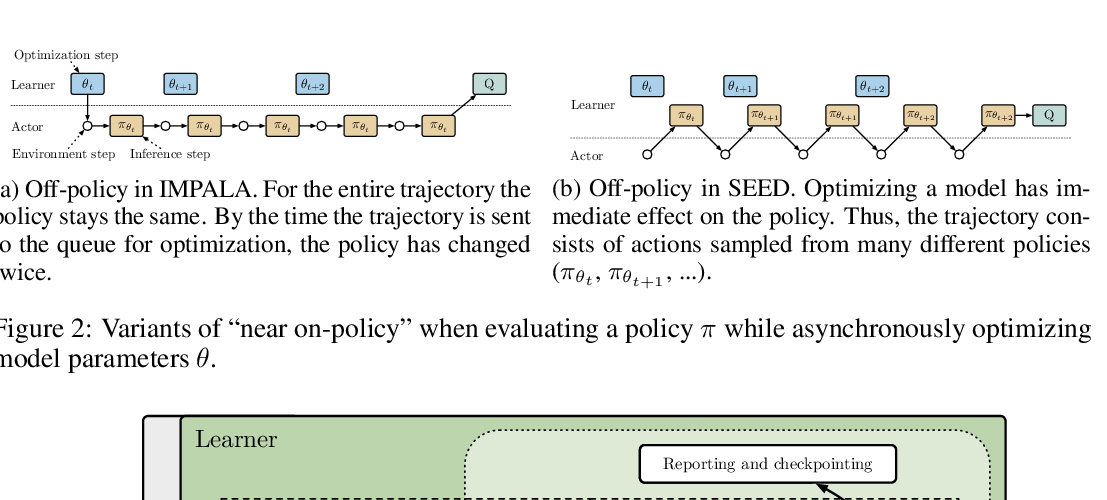
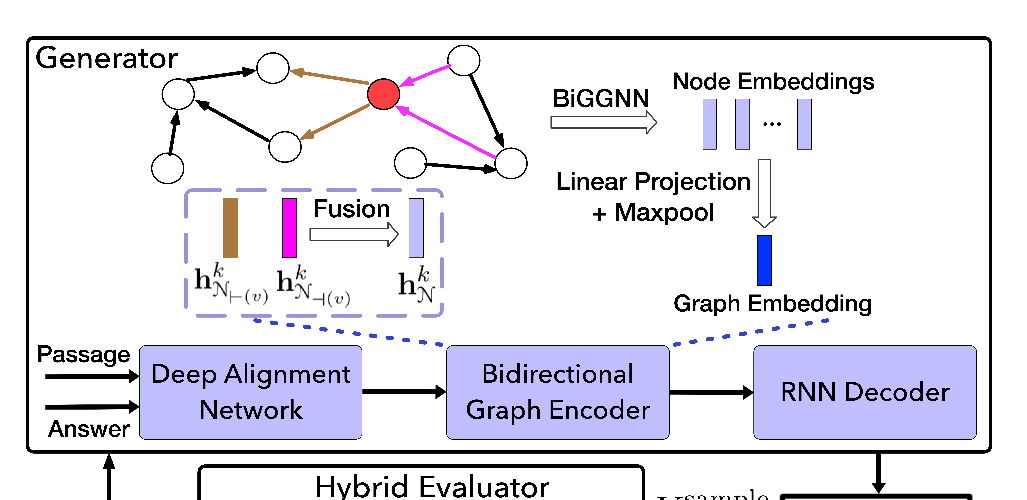
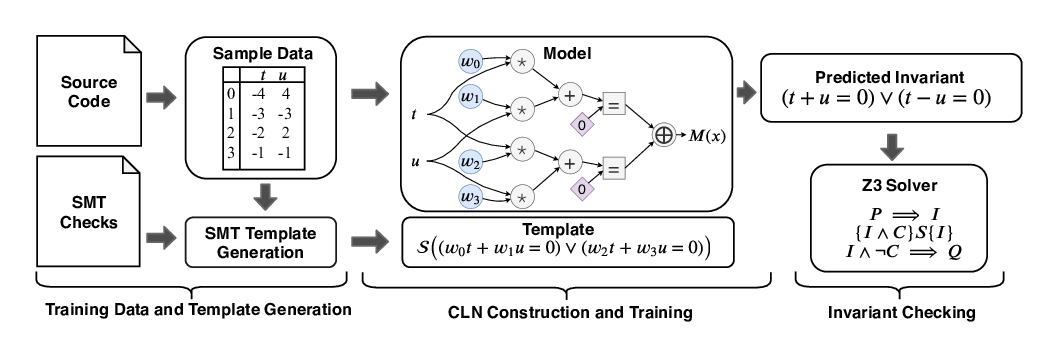
Abstract:
In this paper, we propose an end-to-end deep learning model, called E2Efold, for RNA secondary structure prediction which can effectively take into account the inherent constraints in the problem. The key idea of E2Efold is to directly predict the RNA base-pairing matrix, and use an unrolled algorithm for constrained programming as the template for deep architectures to enforce constraints. With comprehensive experiments on benchmark datasets, we demonstrate the superior performance of E2Efold: it predicts significantly better structures compared to previous SOTA (especially for pseudoknotted structures), while being as efficient as the fastest algorithms in terms of inference time.